This follows the request by Xiamin, Andre and Daren to explore non-active site hits.
NS3 is the protease, its N152–S163 span adopts a beta hairpin that closes up on the substrate when NS2B clamps it down with its own CTD hairpin. Hence NS2B is an activator of NS3.
When there is no NS2B, the span is a disordered loop (open conformation).
Nudge but dont dislodge
Dataset: MatteoFerla-crystal-crackers
For Zika I have previously uploaded the incorrectly named “crystal crackers” set.
These were fiddly semimanual elaborations to expand a non active site pocket to nudge the side the active site, hopefully inhibiting it but not dislodging NS2B (ie.cracking the crystals).
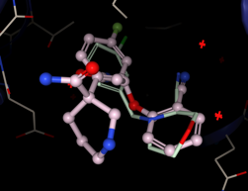
Two hits: both aromatic rings, but angle wrong: reducing ring only solution (Morpholino is more SA than cyclohexane)
Ignore NS2B
Dataset: MatteoFerla-Fragmenstein-offsite-probes
The “non-active site probes” are mergers of the hits 'x0214', 'x0777', 'x0806', 'x0182', 'x0884', 'x0130', 'x0944' in a template simply lacking NS2B and the last few residues of NS3.
I am using the word probe not allosteric modulator as they might simply stick or likely be weaker than NS2B binding.
Hits & their fragments: 56
Virtual hits: 3,751
Shortlist: Dataset sorted by ad hoc metric discussed elsewhere, binned in 20 kmode clusters of interaction sites, fp Tanimoto Butina reduced within each and top 5 of each used.
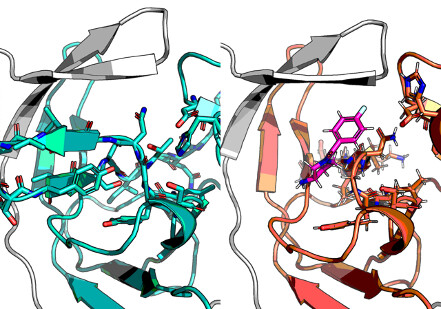
Hypothetical open conformation heterodimer
PyRosetta model: possible albeit weaker than closed.
Designed system, but nothing marvellous came up with OE ROCS in the site otherwise occupied by the closed hairpn
One option is to do in silico mutational scanning (diff in Rosetta pmut_scan between two states) to make a variant that is more stable and test in vivo. But this was not green lit.