The datasets upload to Fragalysis by me for EV D68 3C protease are:
- Manual-Submission
- ROCS&PyRosetta-filtering
- ArthorianQuest-sulfonamide
- Fragmenstein-no_curation
The Manual-Submission are my manual submissions by picking a 3-5 hits from the other three datasets and altering groups if needed, which happened for several. These were ordered.
The Fragmenstein-no_curation was regenerated on Monday 24 July.
It is filtered and sorted but not subjected to human filtering.
This was done with the script fragmenstein_merge_sw_place.py:
python fragmenstein_merge_sw_place.py \
--template reference.pdb \
--hits D68EV3CPROA.corrected.sdf \
--suffix '_v1' \
--sw_databases Enamine-SC-Stock-Mar2022.smi.anon Enamine-BB-Stock-Mar2022.smi.anon REAL-Database-22Q1.smi.anon```
Then in a notebook:
```python
import pandas as pd
from rdkit import Chem
with Chem.SDMolSupplier('D68EV3CPROA.corrected.sdf') as sdf_r:
hits = list(sdf_r)
placements: pd.DataFrame = pd.read_pickle('fragmenstein_placed_v1.pkl.gz')
Added extras
placements['hit_names'] = placements.hit_mols\
.apply(lambda v: v if isinstance(v, list) else [])\
.apply(lambda v: [m.GetProp('_Name') for m in v])
from rdkit import DataStructs
from rdkit.Chem import AllChem
from rdkit.Chem import rdFingerprintGenerator as rdfpg
fpgen = rdfpg.GetRDKitFPGenerator()
hit2fp = {h.GetProp('_Name'): fpgen.GetFingerprint(h) for h in hits}
def get_similarity(row):
if not isinstance(row.minimized_mol, Chem.Mol):
return float('nan')
fp = fpgen.GetFingerprint(AllChem.RemoveHs(row.minimized_mol))
return max([DataStructs.TanimotoSimilarity(fp,hit2fp[name]) for name in row.hit_names])
placements['max_hit_Tanimoto'] = placements.apply(get_similarity, axis=1)
m = placements.minimized_mol.apply(lambda m: m if isinstance(m, Chem.Mol) else Chem.Mol())
placements['largest_ring'] = m.apply(lambda mol: max([0]+ list(map(len, mol.GetRingInfo().AtomRings()))))
import operator
from typing import Dict, List
hit_intxns = pd.read_csv('../D68/hit-intxns.csv', index_col=0)
name2intxns: Dict[str, List[str]] = {'D68EV3CPROA-' + n.split('§')[0]: [(xn.split(':')[0], xn.split(':')[1], int(xn.split(':')[2],)) for xn, xc in xs.items() if xc and xn != 'N_interactions'] for n, xs in hit_intxns.to_dict(orient='index').items()}
def tally_hit_intxns(row: pd.Series):
if not isinstance(row.minimized_mol, Chem.Mol):
return float('nan'), float('nan')
for hit_name in row.hit_names:
present_tally = 0
absent_tally = 0
for intxn_name in name2intxns[hit_name]:
if intxn_name not in row.index:
print(f'missing {intxn_name}')
absent_tally += 1
elif row[intxn_name] == 0:
absent_tally += 1
else:
present_tally += 1
return (present_tally, absent_tally)
hit_checks = placements.apply(tally_hit_intxns, axis=1)
placements['N_interactions_kept'] = hit_checks.apply(operator.itemgetter(0)).fillna(0).astype(int)
placements['N_interactions_lost'] = hit_checks.apply(operator.itemgetter(1)).fillna(99).astype(int)
intxn_names = [c for c in placements.columns if isinstance(c, tuple)]
tallies = placements[intxn_names].sum()
def ratioed(row, k=.5):
return sum([(row[name]/tallies[name])**k for name in intxn_names])
placements['interaction_uniqueness_metric'] = placements.apply(ratioed, axis=1)
import pandas as pd
from rdkit.Chem import PandasTools, Draw
from rdkit import DataStructs
from rdkit.ML.Cluster import Butina
from rdkit.Chem import rdMolDescriptors as rdmd
from rdkit.Chem import Descriptors
from IPython.display import HTML
def butina_cluster(mol_list,cutoff=0.35):
# https://github.com/PatWalters/workshop/blob/master/clustering/taylor_butina.ipynb
fp_list = [rdmd.GetMorganFingerprintAsBitVect(AllChem.RemoveAllHs(m), 3, nBits=2048) for m in mol_list]
dists = []
nfps = len(fp_list)
for i in range(1,nfps):
sims = DataStructs.BulkTanimotoSimilarity(fp_list[i],fp_list[:i])
dists.extend([1-x for x in sims])
mol_clusters = Butina.ClusterData(dists,nfps,cutoff,isDistData=True)
cluster_id_list = [0]*nfps
for idx,cluster in enumerate(mol_clusters,1):
for member in cluster:
cluster_id_list[member] = idx
return cluster_id_list
m = placements.minimized_mol.apply(lambda m: m if isinstance(m, Chem.Mol) else Chem.Mol())
placements['cluster'] = butina_cluster(m.to_list())
import numpy as np
from scipy.spatial import distance
from sklearn.cluster import KMeans
data = placements[[c for c in placements.columns if isinstance(c, tuple)]].values
kmeans = KMeans(n_clusters=50, random_state=42)
kmeans.fit(data)
placements['interaction_cluster'] = kmeans.predict(data)
Finally, I filtered by a weights pulled out of a hat…
import plotly.express as px
weights = {'N_rotatable_bonds': 2,
'∆∆G': +3,
'interaction_uniqueness_metric': -10,
'N_unconstrained_atoms': 1,
'N_constrained_atoms': -0.5,
#'N_HA':
#'LE': -2,
'N_interactions': -5,
'N_interactions_lost': +5, # zero would raise error ...
'max_hit_Tanimoto': -1
}
def weight_split(row):
return [round(row[col] / w, 1) for col, w in weights.items()]
def penalize(row):
penalty = 0
if row.outcome != 'acceptable':
return float('nan')
for col, w in weights.items():
penalty += row[col] * w
return penalty
placements['ad_hoc_penalty'] = placements.apply(penalize, axis=1)
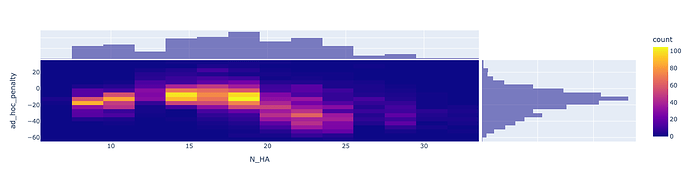
fig = px.density_heatmap(placements.loc[placements.outcome == 'acceptable'],
'N_HA', 'ad_hoc_penalty',
marginal_x="histogram", marginal_y="histogram")
fig.show()
placements\
.loc[placements.outcome == 'acceptable']\
.sort_values('ad_hoc_penalty')\
[['name', 'ad_hoc_penalty', 'smiles', 'N_HA'] + list(weights.keys())]