This post for tag CA - catalytic chain A is here to discuss its contents.
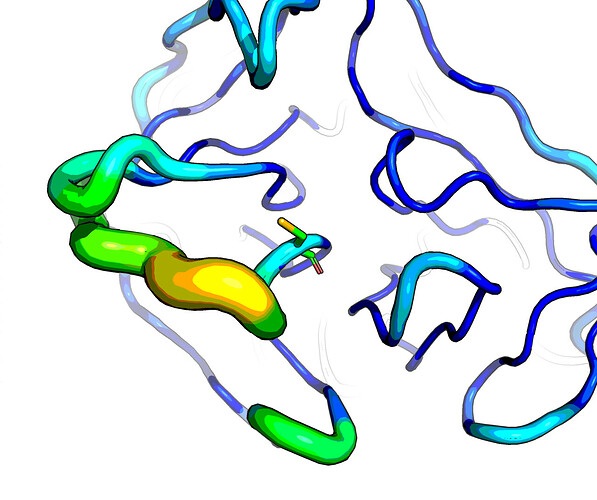
On 28/09/23 there was a discussion on the issue that things do not stick near the catalytic Cys147.
- The covalent library,
CovHetFrags, was used on EV D68 protease, but only hits elsewhere (unreacted warheads) - The oxianion pocket (best viewed in PDB:2B0F) is occupied only in some structures, e.g. x1604_0A, by a water (intxns w/ backbone amide nitrogens of G145 and Q146)
- Two hits interact w/ Cys147 via pi··S:
- x0771_1A (ignore the pyrrolidine & its pucker mode, the benzene)
- X2099_0A (new)
- One via halo-bond:
- 1604_0A (bromo)
- One via H-bond:
- 1594_0A (terminal primary amine)
- Maybe a few others
- Over a dozen hits aimed expanding towards that region were submitted (predictions pictured), but none bound.
- Asn139–Gly145 loop has high B-factors (pictured)
This raises the question of whether this is:
- Chance —Unlikely given other cysteine protease results
- Incompatible solvent — pH8 should mean the cysteine is deprotonated (ie. reactive nucleophile)
- Inaccessible site by soaking —far from simmetry mates
- Not a thermodynamic sink —it’s the catalytic site, i.e. the stabilisation of the oxianion hemiacetal transition state demands it
- Dynamic region in crystals prevents reaction and strong binding
Some solutions were discussed, including mutants.